Crowdsource Computer Game to Help Solve Global Health Problems
Published May-21-12Breakthrough:
An interactive crowdsource game that enables players to help scientists combat diseases by working out how proteins fit together has chalked up numerous success stories - including unlocking the structure of an enzyme that could pave the way for an anti-Aids drug.
Company:
University of Washington, United States
The Story:
 Proteins are the building blocks of life, but they also play a part in many illnesses and conditions. Determining their structure is vital to understand the mechanisms by which diseases are caused and develop, and to create therapeutics to combat them.
Proteins are the building blocks of life, but they also play a part in many illnesses and conditions. Determining their structure is vital to understand the mechanisms by which diseases are caused and develop, and to create therapeutics to combat them.But proteins, even the smallest ones, can be tricky customers. To carry out their tasks they must fold into complex 3D shapes, and they can do this in a huge variety of ways. This makes it a challenge for researchers and their computers to work out their optimum structure.
Scientists know a lot of about the sequence of proteins and their amino acid building blocks, but they know less about how and why these amino acids twist into their intricate shapes. Determining the rules of protein folding remains one of biology’s key challenges.
Mouse Clicking for Medical Science
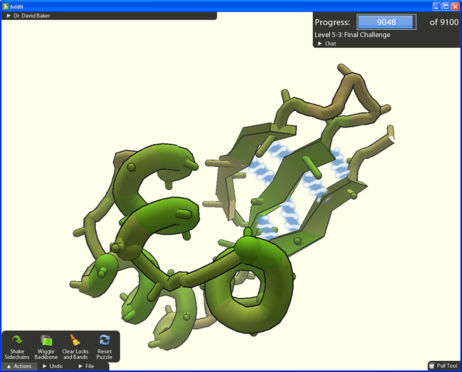
Foldit is an online crowdsource game that encourages players to manipulate digital versions of proteins with a click of their mouse, and in so doing create their optimum folded state. It was developed at the University of Washington by doctoral student Seth Cooper and postdoctoral researcher Adrien Treuille.
The game’s website states that, “Foldit attempts to predict the structure of a protein by taking advantage of humans' puzzle-solving intuitions and having people play competitively to fold the best proteins.”
It's easy to play and a user needs no previous knowledge of biology or chemistry. All that is required is a computer and an Internet connection. Players install a plug-in and then compete with other players to come up with the best structures for proteins.
However, as nobody knows exactly what constitutes the ‘best structures' players are not given any hints toward their goal. They are awarded points not for the structure they create, but by the amount of energy needed to hold in shape a real-life version of the protein.
Groundbreaking Success Story
One particular success story is an enzyme called M-PMV retroviral protease that plays a role in the development of a virus that is similar to HIV. For years scientists had tried, but not succeeded in determining its precise structure. So Foldit players were presented with a digital 3D model of the enzyme which they had to manipulate to try all possible folding combinations. Scientists had spent a decade on the problem; the Foldit crowd solved it in three weeks.
Researchers were so impressed by the results that they included some of the players as co-authors of a study that appeared in the journal Nature: “Crystal structure of a monomeric retroviral protease solved by protein folding game players”.
The crowdsourcing online protein folding game has achieved numerous other milestones including:
Creating an 18-fold more active version of an enzyme that catalyzes the Diels-Alder reaction (used in the synthesis of a number of chemical products).
Harnessing the Brainpower of Gamers
There are more than 100,000 different proteins in the human body and even if you apply all the computing power in the world to understanding protein folding the task will still take centuries. There are just too many possibilities, and so scientists are banking on the intuition of the crowd to get to some of the answers much more quickly.
Next Story »